
In the realm of cancer therapeutics, pharmacogenomics has emerged as a critical linchpin in shaping treatment decisions. The relentless adaptability of cancer cells, often culminating in resistance to chemotherapy and targeted therapies, underscores the pressing need for a more targeted approach. Pharmacogenomics, through the illumination of genetic mutations underpinning drug resistance, offers a powerful compass for oncologists. This genetic insight not only permits the tailoring of individualized treatment plans but also facilitates the precise selection of drugs most likely to exert a therapeutic effect. Furthermore, it spearheads the optimization of drug formulations, honing the delivery mechanisms to their utmost efficacy while minimizing the looming specter of adverse effects. This confluence of genomics and therapeutics not only personalizes cancer treatment but also accelerates the quest for precision medicine, identifying patients poised to reap the greatest benefits from specific treatments, determining the most suitable pharmaceutical agents, and refining drug delivery modalities to enhance treatment outcomes and patient well-being.
Pharmacogenomics and disease
Pharmacogenomics is the science of study of genetic variability and its effects on the drug action. This determines the behavior of a drug inside the individual’s body. Every human being has different genetic makeup due to which there is a difference in the overall metabolic activity so the action of drugs is also expected to be different in individuals which is ultimately affecting the final outcome of the therapy. Applications of pharmacogenomics are limited to the theoretical concept till now and only focused on individualized drug therapy, although if explored to its full potential, it can change the entire therapeutic system with higher degree of accuracy, effectiveness and lowest level of adverse events [1]. Pharmacogenomics may have its applications beyond individualized drug therapy also such as diagnosis of disease, dosage formulation development and evaluation, targeted delivery systems, theranostic delivery systems and polymer activities [2]. Gene’s codes for and control different biological activities (metabolic activities) inside the body via different intermediates such as enzymes, proteins, hormones, and effector cells. These mentioned intermediates may act as the diagnostic and therapeutic biomarkers and may help in diagnosis of disease and designing a suitable dosage form [3]. Identification of genome-based disease specific biomarkers (both diagnostic and therapeutic) is the first step in designing the suitable therapeutic system (fig 1). In the area of cancer research, pharmacogenomics-based approaches of diagnosis and treatment could be of prodigious importance [4]. In cancer therapeutics, pharmacogenomics can be used to guide pharmacotherapy decisions, reduce adverse effects, and improve clinical outcomes. Cancer is a complex disease that involves the growth and spread of abnormal cells in the body. Cancer cells behave differently than the normal cells because variations in receptors, biomolecules and biomarkers (either overexpressed or under expressed), hence, the basic requirements for the cancer cells to be alive is different in comparison to normal cells [5]. The treatment of cancer typically involves a combination of surgery, radiation therapy, and chemotherapy. Chemotherapy drugs work by targeting and killing rapidly dividing cancer cells, but they can also damage healthy cells in the process, leading to side effects. Targeted therapies are drugs that are designed to selectively kill cancer cells by identifying and targeting the specific proteins or pathways that are crucial for tumor growth. However, all patients do not respond to targeted therapies, and some may experience severe side effects [6]. But use of pharmacogenomics-based biomarkers in development of cancer therapy ensures the safety of normal cells via optimization of formulation characteristics as each patient may vary depending on their genetics, age, weight, and other factors so individualized optimization may be helpful [7] (fig. 2). Therefore, along with the guiding pharmacotherapy decisions, pharmacogenomics can also help researchers to develop new cancer drugs that are more effectively and accurately targeted by understanding and identifying the genetic basis of cancer and new drug targets while minimizing damage to healthy cells [8]. Genetic testing can help identify patients who have specific mutations or alterations in genes (diagnostic genomics) that are known to be targeted by the selected and specific drugs (treatment genomics) drugs. By selecting patients who are more likely to respond to treatment, clinicians can improve clinical outcomes and reduce the risk of adverse effects [9]. Genes such as BRCA1, BRCA2, TP53, EGFR, KRAS, HER, ALK, BRAF, PTEN, CDKN2A, RET, MLH1, MSH2, MSH6, PMS2 and VHL are found to have their role in the pathophysiology of different types of cancers (table 1) [10, 11].
Table 1: Genes responsible for induction of various types of cancer
Gene | Role | Type of cancer |
BRCA1 and BRCA2 | Tumor suppression via BRCA1 protein | Mutation increases the risk of developing breast and ovarian cancer |
TP53 | Regulation of cell division | Responsible for breast cancer, colorectal cancer, lung cancer, and many others |
EGFR | Cell surface protein involved in regulation of cell division | Non-small cell lung cancer |
KRAS | Codes for protein involved in cell signalling and regulation of cell division | Indicated for colorectal cancer, lung cancer, and pancreatic cancer |
HER | Codes for a protein involved in regulating cell growth | Over expression is associated with breast cancer |
ALK | Produces protein involved in cell growth and division | Mutations in the ALK gene are responsible for non-small cell lung cancer (NSCLC), anaplastic large-cell lymphoma (ALCL), and neuroblastoma. |
BRAF | Codes for a protein involved in cell signalling and regulation of cell growth | Responsible for melanoma, colorectal cancer, papillary thyroid cancer, and others. |
PTEN | Tumour suppressor gene that codes for a protein involved in regulating cell growth and division | Indicated for breast cancer, prostate cancer, endometrial cancer, and others |
CDKN2A | Tumour suppressor gene that plays a crucial role in regulating the cell cycle | Increased risk of melanoma, pancreatic cancer, and certain types of familial cancer syndromes |
RET | Proto-oncogene that codes for a receptor tyrosine kinase | Risk of Thyroid cancer (papillary thyroid carcinoma), lung cancer (non-small cell lung cancer), and multiple endocrine neoplasia type 2 (MEN2) syndrome |
MLH1 | DNA mismatch repair (MMR) genes | Risk of colorectal cancer, endometrial cancer, ovarian and stomach cancer |
MSH2 / MSH6 | DNA mismatch repair (MMR) genes | Colorectal cancer and endometrial cancer |
PMS2 | DNA mismatch repair (MMR) gene | Risk of colorectal cancer, endometrial cancer, ovarian and stomach cancer |
VHL | Tumour suppressor gene associated with a hereditary cancer syndrome called von Hippel-Lindau disease (VHL). | Responsible for development of various tumours and cysts, particularly in the kidneys, adrenal glands, pancreas, and central nervous system |
Mutation or polymorphism in the genes as mentioned in above table 1, may be responsible for the risk of different types of cancers. Any mutation in these genes can be detected using different techniques such as next generation gene sequencing, multiplex ligation-dependent probe amplification (MLPA), Immunohistochemistry, fluorescence in situ hybridization (FISH) and other. The analysis of the gene mutation can be helpful in understanding the pathophysiology of disease and selection of suitable drug molecules, delivery route, delivery approach and dosage form to manage the cancer in an effective manner.
Pharmacogenomics and drug delivery
Knowledge of pharmacogenomic based interaction of cancer and cancer drugs can help in identification of suitable drug for a specific type of cancer cells and further design and development of a suitable dosage forms which not only delivers the drug in optimal order but also improve the therapeutic outcome of cancer treatment by targeting the drugs to the specific site only. Genes control the synthesis of protein which further control the different physiological mechanisms of body including membrane permeability, transporter proteins, drug carriers, metabolizing enzymes, drug binding proteins, receptor sensitivity, receptor affinity, receptor confirmation and other cellular activities etc. Genes also controls the cellular functions and any alteration in genes may result in elevation or depression of factors (diagnostic biomarkers) that are directly responsible for cancer and involved in its pathophysiology. All these processes collectively affect the pharmacokinetics and pharmacodynamics of drug molecules inside the biological system [12]. Therefore, apart from finding new drug molecules, another approach could be delivering the old molecules with improved and optimized dosage forms. So, dosage formulations may need to be tailored for the individual patients based on their genetic profile. For example, if a patient has a genetic variation that affects how they metabolize a certain drug, a lower dose may be needed to avoid toxicity. Alternatively, a higher dose may be necessary for patients who have a genetic variation that affects how the drug is absorbed or distributed throughout the body [13].
Overall, pharmacogenomics has the potential to improve cancer treatment by providing personalized, more effective and safer therapies and optimized dosage forms as well (Fig. 1).
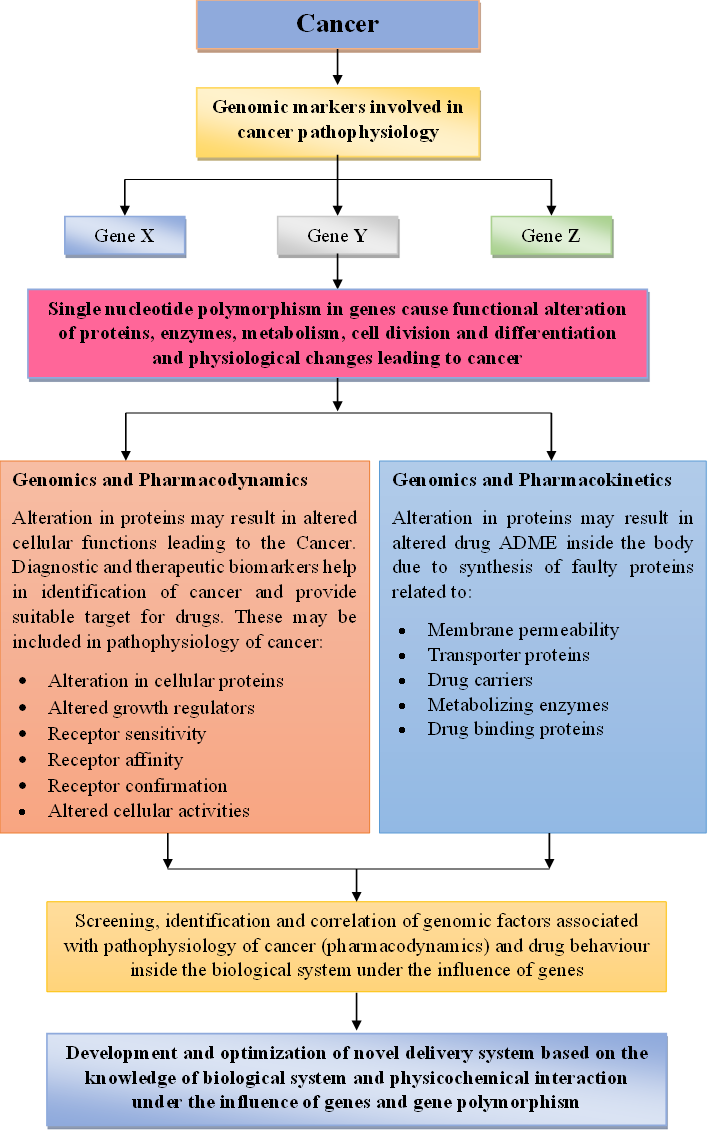
Fig 1: Role of pharmacogenomics in detection of cancer and development of new drugs for treatment

Fig 2: Graphical representation of correlation of cancer and pharmacogenomics in drug delivery optimization
Challenges
Pharmacogenomics, though promising for personalized cancer treatment, faces substantial obstacles. The intricate nature of cancer genetics, with its complex interactions and tumor heterogeneity, makes pinpointing relevant genes difficult. Access to comprehensive genomic profiling is limited by cost and interpretation of vast data requires expertise and sophisticated tools. Additionally, clinical validation of many approaches and uncertainties around reimbursement hinder widespread adoption. Ethical concerns surrounding privacy and discrimination, along with the need for better understanding of gene-drug interactions and integration with clinical workflows, further complicate the landscape. Despite these challenges, ongoing research, technological advancements, and regulatory reforms offer hope for overcoming them and unlocking the true potential of pharmacogenomics in revolutionizing cancer care.
Abbreviations
BRCA1 - Breast Cancer Gene 1
BRCA2 - Breast Cancer Gene 2
TP53 - Tumor protein p53
EGFR - Epidermal Growth Factor Receptor
KRAS - Kirsten rat sarcoma viral oncogene homolog
HER - Human epidermal growth factor receptors
ALK - Anaplastic lymphoma kinase
BRAF - v-Raf murine sarcoma viral oncogene homolog B
PTEN - Phosphatase and Tensin Homolog
CDKN2A - Cyclin-Dependent Kinase Inhibitor 2A
RET - Rearranged During Transfection
MLH1 - MutL Homolog 1
MSH2 - MutS Homolog 2
MSH6 - MutS Homolog 6
PMS2 - Postmeiotic Segregation Increased 2
VHL - von Hippel-Lindau
MLPA - Multiplex ligation-dependent probe amplification
FISH - Fluorescence in situ hybridization
Acknowledgement: None
Author Contribution: All authors contributed equally
Conflict of Interest: Authors declare no conflict of interest
Availability of data and materials: Nil
Funding: Nil
References