
Success in pharmaceutical research can be highly unpredictable. However, more predictive pre-clinical screens can now be implemented to score therapeutic candidates in a way that better correlates with their potential clinical utility. This article explains the influence of predictive validity in analysing such pre-clinical screens to increase productivity in drug discovery.

The macroeconomic fundamentals of the pharmaceutical industry remain strong. High demand for innovative pharmaceutical products is driven by a growing and ageing global human population with significant unmet medical needs. However, discovering new drugs is a costly, lengthy and, still, a largely unpredictable endeavour. The pharmaceutical industry has consistently experienced high rates of compound attrition throughout the drug discovery process over many decades now. Consequently, the biotechnology and pharmaceutical industries are continuing to develop new strategies to improve their overall productivity and reduce the high attrition rates in their development pipelines. Nonetheless, the successful products that do make it on to the market are still having to compensate for too many compounds that are discontinued, as often a lack of compelling efficacy data is uncovered too late during subsequent development. Overall, the number of new drugs approved per billion US dollars spent on research and development has roughly halved about every nine years from 1950 to 2010 with no obvious signs of any substantial improvements over the subsequent decade. So, how can success in pharmaceutical research now become less unpredictable in the future?
Previous analyses have uncovered how assay validity and reproducibility can be correlated across a wide range of simulated screening assays and disease models, such as described in the publication by Scannell and Bosley in 2016 (PLoS ONE 11, 1-21). These authors proposed that increasing the implementation of more relevant and predictive screens should be incorporated pre-clinically into the drug discovery process. A more rigorous understanding of efficacy and toxicity at multiple biological levels would then offer a potential solution to this systemic productivity problem.
Initiating a new drug discovery programme typically requires a chemical starting point for small molecule drugs to initiate the optimisation of that compound’s biological properties, which can also be guided by constantly improving computational chemistry methods. The optimised development candidate needs to possess an adequate balance of efficacy, pharmacokinetics and safety pharmacology to then be progressed into the clinic. Although the discovery process differs somewhat for antibodies and other biological drugs, they are still required to meet adequate biological criteria to be progressed into clinical development. Biological selection criteria need to be driven by assays with higher probability of predicting clinical efficacy in comparison with the current more established workflows. It has been argued that such pivotal decision-making assays need to be introduced much earlier into the discovery process to enable disruptive changes in drug discovery to make a real difference to productivity. For more details on the rationale underlying this conclusion, see the publication by Treherne and Langley in 2021 (Drug Discovery Today 26, 2489-2495).
There have been huge advances in our understanding of the underpinning science of human disease, as well as the introduction of new technologies that should, at least in theoretical principle, have improved the overall productivity of drug discovery. However, success rates still remain stubbornly low. Analytical methods based on decision theory have demonstrated that small changes in the “predictive validity” of an assay have a remarkably significant impact on downstream success rates. In this context, predictive validity refers to the ability of a test or other measurement to predict future outcomes in a human clinical trial. The mathematical basis underlying this approach was described by Scannell et al. in 2022 (Nature Reviews Drug Discovery 21, 915-931), who exemplify the relevance of predictive validity in drug discovery to demonstrate why it matters and then set out how it could be improved. There are many long-standing and unmet medical needs that would benefit from more advanced in vitro assay systems. Historically, drug discovery has been overly dependent on animal models that can be poorly predictive of human pathology, even when human diseased-associated genes are engineered into transgenic mice. Increasingly, more advanced human-specific cellular models are now becoming available. The US Food and Drug Administration (FDA), the US National Institutes of Health and the Environmental Protection Agency have provided long-term support for the incorporation of human cellular models. In fact, animal testing of drugs is no longer required by the FDA following the introduction of a new law that was passed in December 2022. Consequently, the FDA can now approve drugs that have only undergone non-animal testing, such as testing with laboratory-based human tissue models, before proceeding into human clinical trials. The practical implementation of this way of thinking will now be discussed below, particularly in relation to the use of human cellular assays and their more effective exploitation in drug discovery.
In vitro human cellular assays have been well established in drug discovery workflows for the last five decades or more. Explant cultures, for example, are a type of ex vivo tissue culture technique that involves culturing live fragments of human tissues that retain some of the original in vivo characteristics of the source tissue when maintained in culture. Explants can be used in drug discovery to test the efficacy and safety of potential drugs on patient-derived samples and can also be used to study the molecular mechanisms and biomarkers of many diseases. Some advantages of explant cultures are that they can better reflect human biology than most animal models do, as they can preserve the heterogeneity and surrounding microenvironment of the relevant tissue. Some challenges of using explant cultures are that they require a regular supply of fresh, viable tissue samples and they often have limited viability and long-term stability in culture. Cell lines, on the other hand, have been developed to become immortal to provide a reliable and inexhaustible source of human cells. Cell lines were originally used as monolayer or as suspension cultures. However, explant cultures, human tissues and organs are more 3-dimensional (3D). Consequently, spheroid-like cultures have been developed that are typically derived from cell lines that have previously been grown in monolayers or suspensions and then clumped together into 3D-compatible culture systems. Spheroids allow cells to communicate with each other in a similar manner to an in vivo 3D environment, although the lack of vascular flow can still be a limitation. Organoids, on the other hand, are typically referred to as 3D cultures derived from stem cells, which can self-organise in culture owing to their self-renewal and differentiation capacities. Organoids are, typically, seeded and maintained in 3D for the entirety of their life in tissue culture.
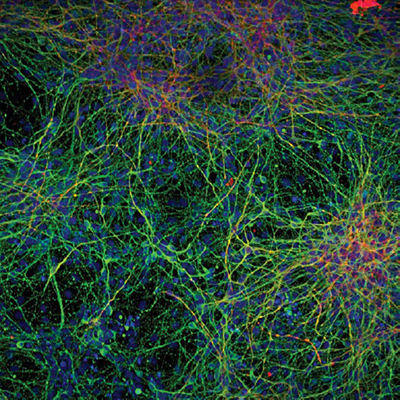
Human induced pluripotent stem cells (iPSCs) can self-renew indefinitely in culture and differentiate into all specialised cell types. Although iPSCs do not exist naturally, they can be “induced” or reprogrammed in culture from any non-reproductive cell. Since they can be generated from any patient with a disease, iPSCs are considered as a valuable technology platform to better model human diseases in vitro. They can then be used to discover new drugs in a variety of screening formats, including the 3D formats described above. For example, iPSCs have been used to develop new screening strategies to treat Alzheimer’s disease by developing assay systems that recapitulate both amyloid and tau pathologies. Such in vitro translational models are now enabling pivotal decisions on compound progression to be made earlier in discovery and can be established in most conventional tissue culture laboratories. The figure below illustrates how iPSs sourced from an individual patient donor can form mature-looking neurons that develop complex morphologies. The image of the neurons shows that Microtubule-Associated Protein 2 (MAP2), which is shown in green, is located predominantly in the neuronal cell bodies and dendrites, whereas β3-tubulin in red is located in the neuronal processes. The neuronal nuclei are shown in blue by staining with DAPI, a fluorescent stain that binds strongly to adenine–thymine-rich regions in DNA. Such complex in vitro cultures are now helping identify drug candidates and new targets for clinical intervention in Alzheimer’s research and other related neurological disorders.
Significant research efforts are also aimed at recapitulating disease models and early-stage efficacy and toxicity screening at the organ level, using in vitro physiological flow systems or “organs-on-chips”. These platforms offer controlled, reproducible and sensitive systems with dynamic flow and tissue-tissue interfaces that support 3D cellular constructs with extended viability. They are amenable to high-content analysis, as they can accommodate electrical, chemical, mechanical and optical sensors and can re-create some aspects of complex human physiology and pathology. Disease modelling in these systems can use human primary cells, conventional cell lines and/or iPSCs. Cells may also be gene edited or subjected to environmental triggers to generate relevant disease pathologies. Unlike simpler cultures, organs-on-chips and fluidically coupled human body-on-chip platforms can give more detailed mechanistic insights into disease processes and the pharmacological effects of compounds. Awareness is needed to understand the limitations of each technology, for example, neuronal cells derived from iPSCs can be relatively immature in some cultures and do not always spontaneously express disease phenotypes. No single platform in isolation is likely to solve the productivity problem in drug development on its own but it is entirely plausible that carefully selected and validated panels of new methodologies could do so.
Developments in organ-on-chip and related cell-based assays continue to support the development of in vitro disease modelling and improved predictions of drug efficacy and toxicity early in drug discovery, leading to the replacement of some animal models. Meta-analyses, comparing in vivo animal toxicity studies with in vitro human-cell high-throughput screening assays, revealed that animal studies did not perform significantly better in predicting adverse drug effects in humans. Both kinds of tests performed only moderately well. However, adding a small set of drug targets to the human-specific in vitro data resulted in models that outperformed those built with the existing animal models.
The costs of introducing new drugs into a practical clinical setting often impedes their extensive use in the patients who most need them. These high costs often arise from the need to compensate for the high attrition rates of potentially promising therapeutics throughout the drug discovery process and into clinical development. Many apparently attractive new drugs fail to deliver meaningful endpoints in clinical trials. This article has analysed the challenges and proposed some novel solutions required to allow the widespread implementation of improved screening strategies into drug discovery. Analytical methods based on decision theory have demonstrated that small changes in the predictive validity of an assay can have a remarkably significant impact on downstream success rates. Predictive validity measurements can then be used to score and rank therapeutic candidates in a way that better correlates with their potential clinical utility. Therefore. Improving the predictive validity of pre-clinical assays in vitro can be used to better predict future outcomes in a human clinical trial. If the productivity problems of the pharmaceutical industry can be overcome by the introduction of these novel screening strategies, then the medical benefits to patients in an ever growing and ageing global population are clear.
References
1. Scannell, JW and Bosley, J. (2016) when quality beats quantity: Decision theory, drug discovery, and the reproducibility crisis. PLoS ONE 11, 1-21
2. Treherne JM and Langley GR (2021) Converging global crises are forcing the rapid adoption of disruptive changes in drug discovery. Drug Discovery Today 26(11), 2489-2495
3. Scannell, JW et al. (2022) Predictive Validity in Drug Discovery: what it is, why it matters, and how to improve it. Nature Reviews Drug Discovery 21(12), 915-931.